Selected publications

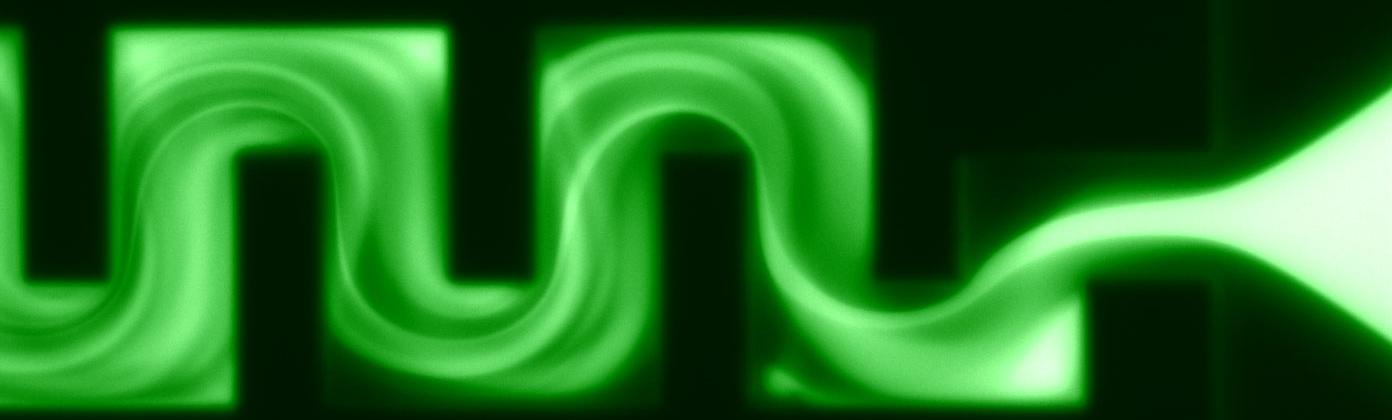
Galvanetto, N., Ivanović, M.T., Chowdhury, A., Sottini, A., Nüesch, M.F., Nettels, D., Best, R.B. & Schuler, B. (2023)
Extreme dynamics in a biomolecular condensate.
Nature 619, 876-883. [PDF]
See News & Views by M. G. Guenza (2023): “Dynamics of protein droplets at multiple scales” Nature 619, 700-701. [PDF]
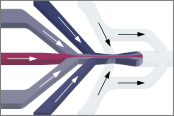
Yang, T., Buholzer, K.J., Sottini, A., Cao, X., deMello, A., Nettels, D. & Schuler, B. (2023)
Rapid droplet-based mixing for single-molecule spectroscopy.
Nat. Methods 20, 1479–1482. [PDF]
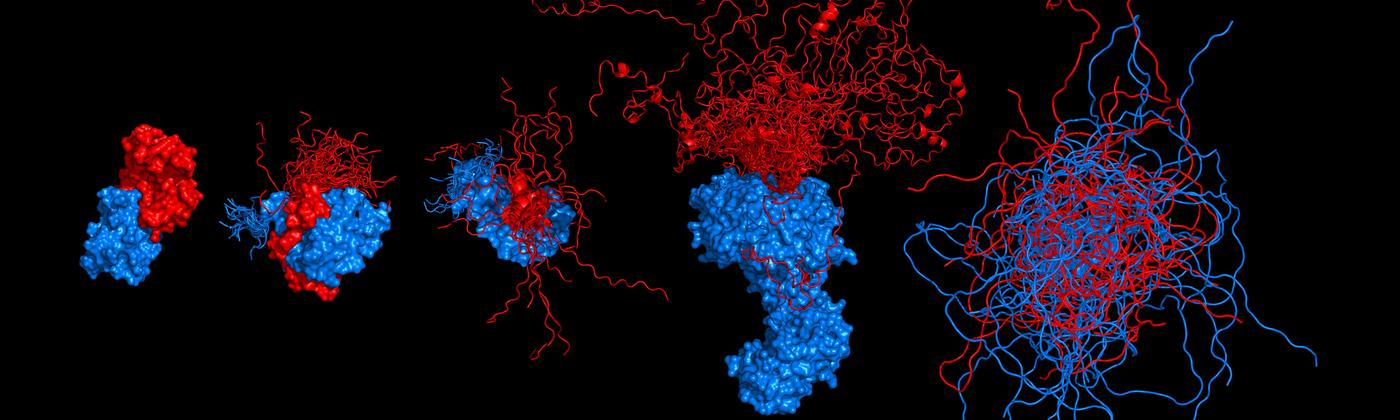
Chowdhury, A., Borgia, A., Ghosh, S., Sottini, A., Mitra, S., Eapen, R.S., Borgia, M. B., Yang, T., Galvanetto, N., Ivanović, M.T., Łukijańczuk, P., Zhu, R., Nettels, D., Kundagrami, A. & Schuler, B. (2023)
Driving forces of the complex formation between highly charged disordered proteins.
Proc. Natl. Acad. Sci. USA 120, e2304036120. [PDF]
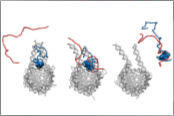
Heidarsson, P.O., Mercadante, D., Sottini, A., Nettels, D., Borgia, M.B., Borgia, A., Kilic, S., Fierz, B, Best, R.B. & Schuler, B. (2022)
Release of linker histone from the nucleosome driven by polyelectrolyte competition with a disordered protein.
Nat. Chem. 14, 224-231. [PDF]
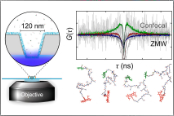
Nüesch, M.F., Ivanovic, M.T., Claude, J.-B., Nettels, D., Best, R.B., Wenger, J. & Schuler, B. (2022)
Single-molecule detection of ultrafast biomolecular dynamics with nanophotonics.
J. Am. Chem. Soc. 144, 52–56. [PDF] ACS Editors’ Choice
See JACS Spotlights “Now You See Me─Refining Detection Methods For Ultra-Fast Proteins” J. Am. Chem. Soc. 144, 1067–1068. [PDF]

Sottini, A., Borgia, A., Borgia, M.B., Bugge, K., Nettels, D., Chowdhury, A., Heidarsson, P.O., Zosel, F., Best, R.B., Kragelund, B.B., & Schuler, B. (2020)
Polyelectrolyte interactions enable rapid association and dissociation in high-affinity disordered protein complexes.
Nat. Commun. 11, 5736. [PDF]
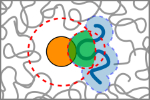
Zosel, F., Soranno, A., Buholzer, K.J., Nettels & Schuler, B. (2020)
Depletion interactions modulate the binding between disordered proteins in crowded environments.
Proc. Natl. Acad. Sci. USA 117, 13480–13489. [PDF]
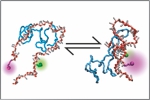
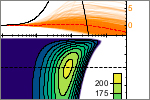

Borgia, A., Borgia, M., Bugge, K., Kissling, V.M., Heidarsson, P.O., Fernandes, C.B., Sottini, A., Soranno, A., Buholzer, K., Nettels, D., Kragelund, B.B., Best, R.B. & Schuler, B. (2018)
Extreme disorder in an ultrahigh-affinity protein complex.
Nature 555, 61-66. [PDF]
See News & Views by Berlow & Wright (2018): “Tight complexes from disordered proteins” Nature 555, 37-38. [PDF]
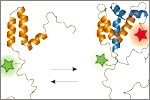
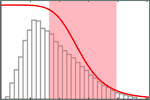
Soranno, A., Holla, A., Dingfelder, F., Nettels, D., Makarov, D.E. & Schuler, B. (2017)
Integrated view of internal friction in unfolded proteins from single-molecule FRET, contact quenching, theory, and simulations.
Proc. Natl. Acad. Sci. USA 114, E1833-E1839. [PDF]

König, I., Zarrine-Afsar, A., Aznauryan, M., Soranno, A., Wunderlich, B., Dingfelder, F., Stüber, J.C., Plückthun, A., Nettels, D. & Schuler, B. (2015)
Single-molecule spectroscopy of protein conformational dynamics in live eukaryotic cells.
Nat. Methods 12, 773-779. [PDF]
See News & Views by Plochowietz & Kapanidis (2015): “Single in the (Cell) City: a proteinfolding story” Nat. Methods 12, 715-716. [PDF]
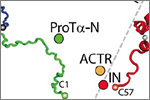
Soranno, A., Koenig, I., Borgia, M.B., Hofmann, H., Zosel, F., Nettels, D. & Schuler, B. (2014)
Single-molecule spectroscopy reveals polymer effects of disordered proteins in crowded environments.
Proc. Natl. Acad. Sci. USA 111, 4874-4879. [PDF]
Wunderlich, B., Nettels, D., Benke, S., Clark, J., Weidner, S., Hofmann, H., Pfeil, S.H. & Schuler, B. (2014)
Microfluidic mixer designed for performing single-molecule kinetics with confocal detection on timescales from milliseconds to minutes.
Nat. Protocols 8, 1459-1474. [PDF]
Soranno, A., Buchli, B., Nettels, D., Cheng, R. R., Müller-Späth, S., Pfeil, S. H., Hoffmann, A., Lipman, E. A., Makarov, D. E. & Schuler, B. (2012)
Quantifying internal friction in unfolded and intrinsically disordered proteins with single-molecule spectroscopy.
Proc. Natl. Acad. Sci. USA 109, 17800–17806. [PDF]
Hofmann, H., Soranno, A., Borgia, A., Gast, K., Nettels, D. & Schuler, B. (2012)
Polymer scaling laws of unfolded and intrinsically disordered proteins quantified with single molecule spectroscopy.
Proc. Natl. Acad. Sci. USA 109, 16155–16160. [PDF]
Borgia, M., Borgia, A., Best, R. B., Steward, A., Nettels, D., Wunderlich, B., Schuler, B. & Clarke, J. (2011)
Single-molecule fluorescence reveals sequence-specific misfolding in multidomain proteins.
Nature 474, 662-665. [PDF]
Müller-Späth, S., Soranno, A., Hirschfeld, V., Hofmann, H., Rüegger, S., Reymond, L., Nettels, D. & Schuler, B. (2010)
Charge interactions can dominate the dimensions of intrinsically disordered proteins.
Proc. Natl. Acad. Sci. USA 107, 14609-14614. [PDF]
See Commentary by England & Haran (2010): “To fold or expand—a charged question” Proc. Natl. Acad. Sci. USA 107, 14519-14520. [PDF]
Recent reviews
Chowdhury, A., Nettels, D. & Schuler, B. (2023)
Interaction Dynamics of Intrinsically Disordered Proteins from Single-Molecule Spectroscopy.
Annu. Rev. Biophys. 52, 433-462. [PDF]
Schuler, B., Borgia, A., Borgia, M.B., Heidarsson, P.O., Holmstrom, E.D., Nettels, D. & Sottini, A. (2020)
Binding without folding – the biomolecular function of disordered polyelectrolyte complexes.
Curr. Opin. Struct. Biol. 60, 66-76. [PDF]
Holmstrom, E.D., Holla, A., Wenwei, Z., Nettels, D., Best, R.B. & Schuler, B. (2018)
Accurate Transfer Efficiencies, Distance Distributions, and Ensembles of Unfolded and Intrinsically Disordered Proteins From Single-Molecule FRET.
Methods Enzymol. 611, 287-325. [PDF]
Schuler, B. (2018)
Perspective: Chain dynamics of unfolded and intrinsically disordered proteins from nanosecond fluorescence correlation spectroscopy combined with single-molecule FRET.
J. Chem. Phys. 149, 010901. [PDF]
Plitzko, J.M., Schuler, B. & Selenko, P. (2017)
Structural Biology outside the box – inside the cell.
Curr. Opin. Struct. Biol. 46, 110–121. [PDF]
Schuler, B., Hofmann, H., Nettels, D. & Soranno, A. (2016)
Single-molecule FRET spectroscopy and the polymer physics of unfolded and intrinsically disordered proteins.
Annu. Rev. Biophys. 45, 207-231. [PDF]
Brucale, M., Schuler, B. & Samori, B. (2014)
Single-Molecule Studies of Intrinsically Disordered Proteins.
Chem. Rev. 114, 3281-3317. [PDF]
Schuler, B. (2013)
Single-molecule FRET of protein structure and dynamics – a primer.
J. Nanobiotechnol. 11 (S1), S2. [PDF]
Schuler, B. & Hofmann, H. (2013)
Single-molecule spectroscopy of protein folding dynamics – expanding scope and timescales.
Curr. Opin. Struct. Biol. 23, 36-47. [PDF]
Complete list of publications
Current Preprints
Nüesch, M.F., Pietrek, L., Holmstrom, E.D., Nettels, D., von Roten, V., Kronenberg-Tenga, R., Medalia, O., Hummer, G. & Schuler, B. (2023)
Nanosecond chain dynamics of single-stranded nucleic acids.
arXiv [PDF]
Grabenhorst, L., Sturzenegger, F., Hasler, M., Schuler, B. & Tinnefeld, P. (2023)
Single-Molecule FRET at 10 MHz Count Rates.
bioRxiv [PDF]
2023
Galvanetto, N., Ivanović, M.T., Chowdhury, A., Sottini, A., Nüesch, M.F., Nettels, D., Best, R.B. & Schuler, B. (2023)
Extreme dynamics in a biomolecular condensate.
Nature 619, 876-883. [PDF]
See News & Views by M. G. Guenza (2023): “Dynamics of protein droplets at multiple scales” Nature 555, 37-38. [PDF]
Yang, T., Buholzer, K.J., Sottini, A., Cao, X., deMello, A., Nettels, D. & Schuler, B. (2023)
Rapid droplet-based mixing for single-molecule spectroscopy.
Nat. Methods 20, 1479–1482. [PDF]
Chowdhury, A., Borgia, A., Ghosh, S., Sottini, A., Mitra, S., Eapen, R.S., Borgia, M. B., Yang, T., Galvanetto, N., Ivanović, M.T., Łukijańczuk, P., Zhu, R., Nettels, D., Kundagrami, A. & Schuler, B. (2023)
Driving forces of the complex formation between highly charged disordered proteins.
Proc. Natl. Acad. Sci. USA 120, e2304036120. [PDF]
Chowdhury, A., Nettels, D. & Schuler, B. (2023)
Interaction Dynamics of Intrinsically Disordered Proteins from Single-Molecule Spectroscopy.
Annu. Rev. Biophys. 52, 433-462. [PDF]
Becker, J., Peters, J.S., Crooks, I., Helmi, S., Synakewicz, M., Schuler, B. & Kukura, P. (2023)
A Quantitative Description for Optical Mass Measurement of Single Biomolecules.
ACS Photonics 10, 2699–2710. [PDF]
2022
Heidarsson, P.O., Mercadante, D., Sottini, A., Nettels, D., Borgia, M.B., Borgia, A., Kilic, S., Fierz, B., Best, R.B. & Schuler, B. (2022)
Release of linker histone from the nucleosome driven by polyelectrolyte competition with a disordered protein.
Nat. Chem. 14, 224-231. [PDF]
Nüesch, M.F., Ivanovic, M.T., Claude, J.-B., Nettels, D., Best, R.B., Wenger, J. & Schuler, B. (2022)
Single-molecule detection of ultrafast biomolecular dynamics with nanophotonics.
J. Am. Chem. Soc. 144, 52–56. [PDF] ACS Editors’ Choice
See JACS Spotlights “Now You See Me─Refining Detection Methods For Ultra-Fast Proteins” J. Am. Chem. Soc. 144, 1067–1068. [PDF]
Buholzer, K.J., McIvor, J., Zosel, F., Teppich, C., Nettels, D., Mercadante, D. & Schuler, B. (2022)
Multilayered allosteric modulation of coupled folding and binding by phosphorylation, peptidyl-prolyl cis/trans isomerization, and diversity of interaction partners.
J. Chem. Phys. 157, 235102. [PDF]
Zosel, F., Holla, A. & Schuler, B. (2022)
Labeling of Proteins for Single-Molecule Fluorescence Spectroscopy.
Protein Folding: Methods and Protocols, Methods Mol. Biol. 2376, 207-233. [PDF]
Stelzl, L.S, Pietrek, L.M., Holla, A., Oroz, J., Sikora, M., Köfinger, J., Schuler, B., Zweckstetter, M., Hummer, G. (2022)
Global Structure of the Intrinsically Disordered Protein Tau Emerges from Its Local Structure.
JACS Au 2, 673–686. [PDF]
Martinsen, J.H., Saar, D., Fernandes, C.B., Schuler, B., Bugge, K., Kragelund, B.B. (2022)
Structure, dynamics, and stability of the globular domain of human linker histone H1.0 and the role of positive charges.
Protein Sci. 31, 918-932. [PDF]
2021
König, I., Soranno, A., Nettels, D. & Schuler, B. (2021)
Impact of in-cell and in-vitro crowding on the conformations and dynamics of an intrinsically disordered protein.
Angew. Chem. Int. Ed. 60, 10724-10729. [PDF]
Benke, S., Holla, A., Wunderlich, B., Soranno, A., Nettels, D. & Schuler, B. (2021)
Combining Rapid Microfluidic Mixing and Three-Color Single-Molecule FRET for Probing the Kinetics of Protein Conformational Changes.
J. Phys. Chem. B, 125, 6617-6628. [PDF]
Dingfelder, F., Macocco, I., Benke, Nettels, D., Faccioli, P. & Schuler, B. (2021)
Slow Escape from a Helical Misfolded State of the Pore-Forming Toxin Cytolysin A.
JACS Au 1, 1217–1230. [PDF]
Klose, D,, Holla, A., Gmeiner, C., Nettels, D., Ritsch, I., Bross, N., Yulikov, M., Allain, F.H.-T., Schuler, B., Jeschke, G. (2021)
Resolving distance variations by single-molecule FRET and EPR spectroscopy using rotamer libraries.
Biophys. J, 120, 4842–4858. [PDF]
See New and Notable by Mingu & Lemke (2021): “When two become one: Integrating FRET and EPR
into one structural model” Biophys. J. 120, 4637–4638. [PDF]
Lerner, E. et al. (2021)
FRET-based dynamic structural biology: Challenges, perspectives and an appeal for open-science practices.
eLife 10, e60416. [PDF]
Stüber, C., Richter, C.P., Bellón, J.S., Schwill, M., König, I., Schuler, B., Piehler, J. & Plückthun, A. (2021)
Apoptosis-inducing anti-HER2 agents operate through oligomerization-induced receptor immobilization.
Commun. Biol. 4, 762. [PDF]
2020
Sottini, A., Borgia, A., Borgia, M.B., Bugge, K., Nettels, D., Chowdhury, A., Heidarsson, P.O., Zosel, F., Best, R.B., Kragelund, B.B. & Schuler, B. (2020)
Polyelectrolyte interactions enable rapid association and dissociation in high-affinity disordered protein complexes.
Nat. Commun. 11, 5736. [PDF]
Zijlstra, N., Nettels, D., Satija, R., Makarov, D.E. & Schuler, B. (2020)
Transition path dynamics of a dielectric particle in a bistable optical trap.
Phys. Rev. Lett. 125, 146001. [PDF]
Zosel, F., Soranno, A., Buholzer, K.J., Nettels, D. & Schuler, B. (2020)
Depletion interactions modulate the binding between disordered proteins in crowded environments.
Proc. Natl. Acad. Sci. USA 117, 13480–13489. [PDF]
Schuler, B., Borgia, A., Borgia, M.B., Heidarsson, P.O., Holmstrom, E.D., Nettels, D. & Sottini, A. (2020)
Binding without folding – the biomolecular function of disordered polyelectrolyte complexes.
Curr. Opin. Struct. Biol. 60, 66-76. [PDF]
Gosavi, S. & Schuler, B. (2020)
Editorial Overview: Molecular interactions that drive folding and binding: new challenges and opportunities.
Curr. Opin. Struct. Biol. 60, iii-iv. [PDF]
Ernst, P., Zosel, F., Reichen, C., Nettels, D., Schuler, B. & Plückthun, A. (2020)
Structure-guided design of a peptide lock for modular peptide binders.
ACS Chem. Biol. 2020, 15, 457-468. [PDF]
2019
Holmstrom, E.D., Liu, Z., Nettels, D., Best, R.B. & Schuler, B. (2019)
Disordered RNA chaperones can enhance nucleic acid folding via local charge screening.
Nat. Commun. 10, 2453. [PDF]
Assenza, S., Sassi, A.S., Kellner, R., Schuler, B., De Los Rios, P. & Barducci, A. (2019)
Efficient conversion of chemical energy into mechanical work by Hsp70 chaperones.
eLife 8:e48491. [PDF]
2018
Borgia, A., Borgia, M., Bugge, K., Kissling, V.M., Heidarsson, P.O., Fernandes, C.B., Sottini, A., Soranno, A., Buholzer, K., Nettels, D., Kragelund, B.B., Best, R.B. & Schuler, B. (2018)
Extreme disorder in an ultrahigh-affinity protein complex.
Nature 555, 61-66. [PDF]
See News & Views by Berlow & Wright (2018): “Tight complexes from disordered proteins” Nature 555, 37-38. [PDF]
Sturzenegger, F., Zosel, F., Holmstrom, E.D., Buholzer, K.J, Makarov, D.E., Nettels, D. & Schuler, B. (2018)
Transition path times of coupled folding and binding reveal the formation of an encounter complex.
Nat. Commun. 9, 4708. [PDF]
Zosel, F., Mercadante, D., Nettels, D. & Schuler, B. (2018)
A proline switch explains kinetic heterogeneity in a coupled folding and binding reaction.
Nat. Commun. 9, 3332. [PDF]
Dingfelder, F., Benke, S., Nettels, D. & Schuler B. (2018)
Mapping an Equilibrium Folding Intermediate of the Cytolytic Pore Toxin ClyA with Single-Molecule FRET.
J. Phys. Chem. B 122, 11251-11261. (Festschrift on the occasion of the 80th birthday of William A. Eaton) [PDF]
Zheng, W., Hofmann, H., Schuler, B. & Best, R.B. (2018)
Origin of Internal Friction in Disordered Proteins Depends on Solvent Quality.
J. Phys. Chem. B 122, 11478-11487. [PDF]
Grotz, K.K., Nueesch, M.F., Holmstrom, E.D., Heinz, M., Stelzl, L.S., Schuler, B. & Hummer, G. (2018)
Dispersion Correction Alleviates Dye Stacking of Single-Stranded DNA and RNA in Simulations of Single-Molecule Fluorescence Experiments.
J. Phys. Chem. B 122, 11626-11639. [PDF]
Marino, J., Buholzer, K.J., Zosel, F., Nettels, D. & Schuler, B. (2018)
Charge Interactions Can Dominate Coupled Folding and Binding on the Ribosome.
Biophys J. 115, 996-1006. [PDF]
Zheng, W., Zerze, G.H., Borgia, A., Mittal, J., Schuler, B. & Best, R.B. (2018)
Inferring properties of disordered chains from FRET transfer efficiencies.
J. Chem. Phys. 148, 123329. (Special Issue on Single-Molecule Biophysics) [PDF]
Holmstrom, E.D., Holla, A., Wenwei, Z., Nettels, D., Best, R.B. & Schuler, B. (2018)
Accurate Transfer Efficiencies, Distance Distributions, and Ensembles of Unfolded and Intrinsically Disordered Proteins From Single-Molecule FRET.
Methods Enzymol. 611, 287-325. [PDF]
Best, R.B., Zheng, W., Borgia, A., Buholzer, K., Borgia, M.B., Hofmann, H., Soranno, A., Nettels. D., Gast, K., Grishaev, A. & Schuler, B. (2018)
Comment on “Innovative scattering analysis shows that hydrophobic disordered proteins are expanded in water”.
Science 361, eaar7101. [PDF]
Hellenkamp, B., Schmid, S., Doroshenko, O., Opanasyuk, O., Kühnemuth, R., Adariani, S.R., Ambrose, B., Aznauryan, M., Barth, A., Birkedal, V., Bowen, M.E., Chen, H., Cordes, T., Eilert, T., Fijen, C., Gebhardt, Ch., Götz, M., Gouridis, G., Gratton, E., Ha, T., Hao, P., Hanke, Ch.A., Hartmann, A., Hendrix, J., Hildebrandt, L.L., Hirschfeld, V., Hohlbein, J., Hua, B., Hübner, Ch.G, Kallis, E., Kapanidis, A.N., Kim, J.Y., Krainer, G., Lamb, D.C., Lee, N.K., Lemke, E.A., Levesque, B., Levitus, M., McCann, J.J, Naredi-Rainer, N., Nettels, D., Ngo, T., Qiu, R., Robb, N.C., Röcker, C., Sanabria, H., Schlierf, M., Schröder, T., Schuler, B., Seidel, H., Streit, L., Thurn, J., Tinnefeld, Ph., Tyagi, S., Vandenberk, N., Vera, A.M., Weninger, K.R., Wünsch, B., Yanez-Orozco, I.S., Michaelis, J., Seidel, C.A.M., Craggs, T.D., & Hugel, T. (2018)
Precision and accuracy of single-molecule FRET measurements—a multi-laboratory benchmark study.
Nat. Methods 15, 669-676. [PDF]
Schuler, B. (2018)
Perspective: Chain dynamics of unfolded and intrinsically disordered proteins from nanosecond fluorescence correlation spectroscopy combined with single-molecule FRET.
J. Chem. Phys. 149, 010901. [PDF]
Nasrollah, R.-G., Parigi, G., Soranno, A., Holla, A., Becker, S., Schuler, B., Luchinat, C. & Zweckstetter, M. (2018)
Local and Global Dynamics in Intrinsically Disordered Synuclein.
Angew. Chem. Int. Ed. 57, 15262 –15266. [PDF]
Angew. Chem. 130, 15482-15486. [PDF]
Holmstrom, E., Nettels, D. & Schuler, B. (2018)
Conformational Plasticity of Hepatitis C Virus Core Protein Enables RNA-Induced Formation of Nucleocapsid-like Particles.
J. Mol. Biol. 430, 2453-2467. (Special Issue on Intrinsically Disordered Proteins) [PDF]
Reinartz, I., Sinner, C., Nettels, D., Stucki-Buchli, B., Stockmar, F., Panek, P.T., Jacob, C.R., Nienhaus, G.U., Schuler, B. & Schug, A. (2018)
Simulation of FRET Dyes Allows Quantitative Comparison against Experimental Data.
J. Chem. Phys. 148, 123321. (Special Issue on Single-Molecule Biophysics) [PDF]
Makarov, D. E. & Schuler, B. (2018)
Preface: Special Topic on Single-Molecule Biophysics.
J. Chem. Phys. 148, 123001. [PDF]
See https://aip.scitation.org/toc/jcp/148/12 for the complete Special Topic.
Masliah, G., Maris, C., König, S.L.B., Yulikov, M., Aeschimann, F., Mabille, J., Weiler, J., Holla, A., Hunziker, J., Meisner-Kober, N., Schuler, B., Jeschke, G. & Allain, F.H.-T. (2018)
Structural basis of siRNA recognition by TRBP double-stranded RNA binding domains.
EMBO J. 37, e97089. [PDF]
Hansen, S., Ernst, P., König, S.L.B., Reichen, C., Ewald, C., Nettels, D., Mittl, P.R.E., Schuler, B. & Plückthun, A. (2018)
Curvature of designed armadillo repeat proteins allows modular peptide binding.
J. Struct. Biol. 201, 108-117. (Special Issue on Proteins with Tandem Repeats) [PDF]
Morger, D., Zosel, F., Bühlmann, M., Züger, S., Mittelviefhaus, M., Schuler, B., Luban, J. & Grütter, M.G. (2018)
The three-fold axis of the HIV-1 capsid lattice is the species-specific binding interface for TRIM5α.
J. Virol. 92, e01541-17. [PDF]
2017
Soranno, A., Holla, A., Dingfelder, F., Nettels, D., Makarov, D.E. & Schuler, B. (2017)
Integrated view of internal friction in unfolded proteins from single-molecule FRET, contact quenching, theory, and simulations.
Proc. Natl. Acad. Sci. USA 114, E1833-E1839. [PDF]
Zijlstra, N., Dingfelder, F., Wunderlich, B., Zosel, F., Benke, S., Nettels, D., & Schuler, B. (2017)
Rapid Microfluidic Dilution for Single-Molecule Spectroscopy of Low-Affinity Biomolecular Complexes.
Angew. Chem. Int. Ed. 56, 7126–7129. [PDF]
Dingfelder, F., Wunderlich, B., Benke, S., Zosel, F., Zijlstra, N., Nettels, D. & Schuler, B. (2017)
Rapid Microfluidic Double-Jump Mixing Device for Single-Molecule Spectroscopy.
J. Am. Chem. Soc. 139, 6062-6065. [PDF]
Plitzko, J.M., Schuler, B. & Selenko, P. (2017)
Structural Biology outside the box — inside the cell.
Curr. Opin. Struct. Biol. 46, 110–121. [PDF]
Zosel, F., Haenni, D., Soranno, A., Nettels, D. & Schuler, B. (2017)
Combining short- and long-range fluorescence reporters with simulations to explore the intramolecular dynamics of an intrinsically disordered protein.
J. Chem. Phys. 147, 152708. (Special Topic Issue: Reaction Pathways) [PDF]
Benke, S., Nettels, D., Hofmann, H. & Schuler, B. (2017)
Quantifying kinetics from time series of single-molecule Förster resonance energy transfer efficiency histograms.
Nanotechnology 28, 114002. (Focus Collection on Protein Folding) [PDF]
Ruggeri, F., Zosel, F., Mutter, N., Różycka, M., Wojtas, M., Ożyhar, A., Schuler, B. & Krishnan, M. (2017)
Single-molecule electrometry.
Nat. Nanotechnol. 12, 488-495. [PDF]
2016
Aznauryan, M., Delgado, L., Soranno, A., Nettels, D., Huang, J., Labhardt, A.M., Grzesiek, S. & Schuler, B. (2016)
Comprehensive structural and dynamical view of an unfolded protein from the combination of single-molecule FRET, NMR and SAXS.
Proc. Natl. Acad. Sci. USA 113, E5389–E5398. [PDF]
Borgia, A., Zheng W., Buholzer, K., Borgia, M., Schüler A., Hofmann, H., Soranno, A., Nettels, D., Gast, K., Grishaev, A., Best, R.B. & Schuler, B. (2016)
Consistent View of Polypeptide Chain Expansion in Chemical Denaturants from Multiple Experimental Methods.
J. Am. Chem. Soc. 138, 11714−11726. [PDF]
See JACS Spotlight “Settling a Folding Debate Once and For All”, J. Am. Chem. Soc. 2016, 138, 13083−13084. [PDF]
Zheng, W., Borgia, A., Buholzer, K., Grishaev, A., Schuler, B. & Best., R.B. (2016)
Probing the Action of Chemical Denaturant on an Intrinsically Disordered Protein by Simulation and Experiment.
J. Am. Chem. Soc. 138, 11702−11713. [PDF]
Schuler, B., Hofmann, H., Nettels, D. & Soranno, A. (2016)
Single-molecule FRET spectroscopy and the polymer physics of unfolded and intrinsically disordered proteins.
Annu. Rev. Biophys. 45, 207-231. [PDF]
Roderer, D., Benke, S., Schuler, B. & Glockshuber, R. (2016)
Soluble Oligomers of the Pore-forming Toxin Cytolysin A from Escherichia coli Are Off-pathway Products of Pore Assembly.
J. Biol. Chem. 291, 5652–5663. [PDF]
2015
König, I., Zarrine-Afsar, A., Aznauryan, M., Soranno, A., Wunderlich, B., Dingfelder, F., Stüber, J.C., Plückthun, A., Nettels, D. & Schuler, B. (2015)
Single-molecule spectroscopy of protein conformational dynamics in live eukaryotic cells.
Nat. Methods 12, 773-779. [PDF]
See News & Views by Plochowietz & Kapanidis (2015): “Single in the (Cell) City: a protein-folding story” Nat. Methods 12, 715-716. [PDF]
Borgia, A., Kemplen, K.R., Borgia, M.B., Soranno, A., Shammas, S., Wunderlich, B., Nettels, D., Best, R.B., Clarke, J. & Schuler, B. (2015)
Transient misfolding dominates multidomain protein folding.
Nat. Commun. 6, 8861. [PDF]
Benke, S., Roderer, D., Wunderlich, B., Nettels, D., Glockshuber, R. & Schuler, B. (2015)
The assembly dynamics of the cytolytic pore toxin ClyA.
Nat. Commun. 6, 6198. [PDF]
Nettels, D., Haenni, D., Maillot, S., Gueye, M., Barth, A., Hirschfeld, V., Hübner, C.G., Léonard, J. & Schuler, B. (2015)
Excited-state annihilation reduces power dependence of single-molecule FRET experiments.
Phys. Chem. Chem. Phys. 17, 32304-32315. [PDF]
Best, R.B., Hofmann, H., Nettels, D. & Schuler, B. (2015)
Quantitative Interpretation of FRET Experiments via Molecular Simulation: Force Field and Validation.
Biophys. J. 108, 2721-2731. [PDF]
Zheng, W., Borgia, A., Borgia, M., Schuler, B. & Best, R.B. (2015)
Empirical Optimization of Interactions between Proteins and Chemical Denaturants in Molecular Simulations.
J. Chem. Theory Comput. 11, 5543-5553. [PDF]
Schuler, B. & Smith, J.L. (2015)
Editorial overview: Biophysical and molecular biological methods: Structure, dynamics, and single molecules.
Curr. Opin. Struct. Biol. 34, iv-vi. [PDF]
Czar, M.F., Zosel, F., König, I., Nettels, D., Wunderlich, B., Schuler, B., Zarrine-Afsar, A. & Jockusch, RA. (2015)
Gas-Phase FRET Efficiency Measurements To Probe the Conformation of Mass-Selected Proteins.
Anal. Chem. 87, 7559-7565. [PDF]
Yuan, H., Gaiduk, A., Siekierzycka, J.R., Fujiyoshi, S., Matsushita, M., Nettels, D., Schuler, B., Seidel, C.A. & Orrit, M. (2015)
Temperature-cycle microscopy reveals single-molecule conformational heterogeneity.
Phys. Chem. Chem. Phys. 17, 6532-6544. [PDF]
2014
Soranno, A., Koenig, I., Borgia, M.B., Hofmann, H., Zosel, F., Nettels, D. & Schuler, B. (2014)
Single-molecule spectroscopy reveals polymer effects of disordered proteins in crowded environments.
Proc. Natl. Acad. Sci. USA 111, 4874-4879. [PDF]
Wuttke, R., Hofmann, H., Nettels, D., Borgia, M.B., Mittal, J., Best, R.B. & Schuler, B. (2014)
Temperature-dependent solvation modulates the dimensions of disordered proteins.
Proc. Natl. Acad. Sci. USA 111, 5213–5218. [PDF]
Kellner, R., Hofmann, H., Barducci, A., Wunderlich, B., Nettels, D. & Schuler, B. (2014)
Single-molecule spectroscopy reveals chaperone-mediated expansion of substrate protein.
Proc. Natl. Acad. Sci. USA 111, 13355-13360. [PDF]
Wunderlich, B., Nettels, D. & Schuler, B. (2014)
Taylor dispersion and the position-to-time conversion in microfluidic mixing devices.
Lab. Chip 14, 219-228. [PDF]
Hofmann, H., Hillger, F., Delley, C., Hoffmann, A., Pfeil, S.H., Nettels, D., Lipman, E.A. & Schuler, B. (2014)
Role of denatured-state properties in chaperonin action probed by single-molecule spectroscopy.
Biophys. J. 107, 2891-2902. [PDF]
Pochorovski, I., Knehans, T., Nettels, D., Müller, A.M., Schweizer, W.B., Caflisch, A., Schuler, B. & Diederich, F. (2014)
Experimental and Computational Study of BODIPY Dye-Labeled Cavitand Dynamics.
J. Am. Chem. Soc. 136, 2441-2449. [PDF]
Brucale, M., Schuler, B. & Samori, B. (2014)
Single-Molecule Studies of Intrinsically Disordered Proteins.
Chem. Rev. 114, 3281-3317. [PDF]
Weisenburger, S., Jing, B., Hänni, D., Reymond, L., Schuler, B., Renn, A. & Sandoghdar, V. (2014)
Cryogenic Colocalization Microscopy for Nanometer-Distance Measurements.
ChemPhysChem 15, 763-770. [PDF]
Roderer, D., Benke, S., Müller, M., Fäh-Rechsteiner, H., Ban, N., Schuler, B. & Glockshuber, R (2014)
Characterization of Variants of the Pore-Forming Toxin ClyA from Escherichia coli Controlled by a Redox Switch.
Biochemistry 53, 6357-6369. [PDF]
2013
Wunderlich, B., Nettels, D., Benke, S., Clark, J., Weidner, S., Hofmann, H., Pfeil, S.H. & Schuler, B. (2013)
Microfluidic mixer designed for performing single-molecule kinetics with confocal detection on timescales from milliseconds to minutes.
Nat. Protocols 8, 1459-1474. [PDF]
Aznauryan, M., Nettels, D., Holla, A., Hofmann, H. & Schuler, B. (2013)
Single-molecule spectroscopy of cold denaturation and the temperature-induced collapse of unfolded proteins.
J. Am. Chem. Soc. 135, 14040-14043. [PDF]
Haenni, D., Zosel, F., Reymond, L., Nettels, D. & Schuler, B. (2013)
Intramolecular distances and dynamics from the combined photon statistics of single-molecule FRET and photoinduced electron transfer.
J. Phys. Chem. B 42, 13015-28. (Festschrift on the occasion of the 60th birthday of Peter W. Wolynes) [PDF]
Hofmann, H., Nettels, D. & Schuler, B. (2013)
Single-molecule spectroscopy of the unexpected collapse of an unfolded protein at low pH.
J. Chem. Phys. 139, 121930. (Special Issue: Chemical Physics of Biological Systems) [PDF]
Schuler, B. & Clarke, J. (2013)
Rough passage across a barrier.
Nature 502, 632-633. [PDF]
Schuler, B. (2013)
Single-molecule FRET of protein structure and dynamics – a primer.
J. Nanobiotechnol. 11 (S1), S2. [PDF]
Schuler, B. & Hofmann, H. (2013)
Single-molecule spectroscopy of protein folding dynamics-expanding scope and timescales.
Curr. Opin. Struct. Biol. 23, 36-47. [PDF]
2012
Hofmann, H., Soranno, A., Borgia, A., Gast, K., Nettels, D. & Schuler, B. (2012)
Polymer scaling laws of unfolded and intrinsically disordered proteins quantified with single molecule spectroscopy.
Proc. Natl. Acad. Sci. USA 109, 16155–16160. [PDF]
Soranno, A., Buchli, B., Nettels, D., Cheng, R. R., Müller-Späth, S., Pfeil, S. H., Hoffmann, A., Lipman, E. A., Makarov, D. E. & Schuler, B. (2012)
Quantifying internal friction in unfolded and intrinsically disordered proteins with single-molecule spectroscopy.
Proc. Natl. Acad. Sci. USA 109, 17800–17806. [PDF]
Borgia, A., Wensley, B.G., Soranno, A., Nettels, D., Borgia, M.B., Hoffmann, A., Pfeil, S.H., Lipman, E.A., Clarke, J. & Schuler, B. (2012)
Localizing internal friction along the reaction coordinate of protein folding by combining ensemble and single-molecule fluorescence spectroscopy.
Nat. Commun. 3, 1195. [PDF]
2011
Borgia, M., Borgia, A., Best, R. B., Steward, A., Nettels, D., Wunderlich, B., Schuler, B. & Clarke, J. (2011)
Single-molecule fluorescence reveals sequence-specific misfolding in multidomain proteins.
Nature 474, 662-665. [PDF]
Hoffmann, A., Nettels, D., Clark, J., Borgia, A., Radford, S.E., Clarke, J. & Schuler, B. (2011)
Quantifying heterogeneity and conformational dynamics from single molecule FRET of diffusing molecules: Recurrence analysis of single particles (RASP).
PhysChemChemPhys 13, 1857-1871. (Special issue on Single-Molecule Optical Studies of Soft and Complex Matter) [PDF]
Hoefling, M., Lima, N., Haenni, D., Seidel, CA., Schuler, B. & Grubmüller, H. (2011)
Structural heterogeneity and quantitative FRET efficiency distributions of polyprolines through a hybrid atomistic simulation and Monte Carlo approach.
PLOS ONE 6, e19791. [PDF]
Yuan, H., Xia, T., Schuler, B. & Orrit, M. (2011)
Temperature-cycle single-molecule FRET microscopy on polyprolines.
PhysChemChemPhys 13, 1762-1769. (Special issue on Single-Molecule Optical Studies of Soft and Complex Matter) [PDF]
2010
Müller-Späth, S., Soranno, A., Hirschfeld, V., Hofmann, H., Rüegger, S., Reymond, L., Nettels, D. & Schuler, B. (2010)
Charge interactions can dominate the dimensions of intrinsically disordered proteins.
Proc. Natl. Acad. Sci. USA 107, 14609-14614. [PDF]
See Commentary by England & Haran (2010): “To fold or expand—a charged question” Proc. Natl. Acad. Sci. USA 107, 14519-14520. [PDF]
Hofmann, H., Hillger, F., Pfeil, S. H., Hoffmann, A., Streich, D., Haenni, D., Nettels, D., Lipman, E. A. & Schuler, B. (2010)
Single-molecule spectroscopy of protein folding in a chaperonin cage.
Proc. Natl. Acad. Sci. USA 107, 11793-11798. [PDF]
Schuetz, P., Wuttke, R., Schuler, B. & Caflisch, A. (2010)
Free Energy Surfaces from Single-Distance Information.
J. Phys. Chem. B 114, 15227–15235. [PDF]
2009
Nettels, D., Müller-Späth, S., Küster, F., Hofmann, H., Haenni, D., Rüegger, S., Reymond, L., Hoffmann, A., Kubelka, J., Heinz, B., Gast, K., Best, R.B. & Schuler, B. (2009)
Single molecule spectroscopy of the temperature-induced collapse of unfolded proteins.
Proc. Natl. Acad. Sci. USA 106, 20740-20745. [PDF]
Gopich, I.V., Nettels, D., Schuler, B. & Szabo, A. (2009)
Protein dynamics from single-molecule intensity correlation functions.
J. Chem. Phys. 131, 095102. [PDF]
2008
Hillger, F., Hänni, D., Nettels, D., Geister, S., Grandin, M., Textor, M. & Schuler, B. (2008)
Probing protein-chaperone interactions with single molecule fluorescence spectroscopy.
Angew. Chem. Int. Ed. 47, 6184-6188. [PDF]
Angew. Chem. 120, 6183-6194. [PDF]
Nettels, D., Hoffmann, A. & Schuler, B. (2008)
Unfolded protein and peptide dynamics investigated with single molecule FRET and correlation spectroscopy from picoseconds to seconds.
J. Phys. Chem. B 112, 6137-6146. (Festschrift on the occasion of the 60th birthday of Attila Szabo) [PDF]
Wahl, M, Rahn, H.-J., Röhlicke, T., Kell, G., Nettels, D., Hillger, F., Schuler, B. & Erdmann, R. (2008)
Scalable time-correlated photon counting system with multiple independent input channels.
Rev. Sci. Instrum. 79, 123113. [PDF]
Schuler, B. & Eaton, W. A. (2008)
Protein folding studied by single molecule FRET.
Curr. Opin. Struct. Biol. 18, 16-26. [PDF]
Kane, A. S., Hoffmann, A., Baumgärtel, P., Seckler, R., Reichardt, G., Horsley, D. A., Schuler, B. & Bakajin, O. (2008)
Microfluidic Mixers for the Investigation of Rapid Protein Folding Kinetics Using Synchrotron Radiation Circular Dichroism Spectroscopy.
Anal. Chem. 80, 9534-9541. [PDF]
2007
Nettels, D., Gopich, V.I., Hoffmann, A. & Schuler, B. (2007)
Ultrafast dynamics of protein collapse from single molecule photon statistics.
Proc. Natl. Acad. Sci. USA 104, 2655-2660. [PDF]
See Highlight in Science 315, 1194, “Editors’ choice”.
Hoffmann, A., Kane, A., Nettels, D., Hertzog, D., Baumgärtel, P., Lengefeld, J., Reichardt, G., Horsley, D.A., Seckler, R., Bakajin, O. & Schuler, B. (2007)
Mapping protein collapse with single molecule fluorescence and kinetic synchrotron radiation circular dichroism spectroscopy.
Proc. Natl. Acad. Sci. USA 104, 105-110. [PDF]
Nettels, D. & Schuler, B. (2007)
Subpopulation-resolved photon statistics of single-molecule energy transfer dynamics.
IEEE J. Sel. Top. Quant. 313, 990-995. [PDF]
Hillger, F., Nettels, D., Dorsch, S. & Schuler, B. (2007)
Detection and analysis of protein aggregation with confocal single molecule fluorescence spectroscopy.
J. Fluoresc. (Special Issue: Single Molecule Spectroscopy) 17, 759-765. [PDF]
Schuler, B. (2007)
Application of Single Molecule Förster Resonance Energy Transfer to Protein Folding.
Protein Folding Protocols (Humana Press; Bai, Nussinov, Eds.), 115–138. [PDF]
Best, R.B., Merchant, K.A., Gopich, I.V., Schuler, B., Bax, A. & Eaton, W.A. (2007)
Effect of flexibility and cis residues in single-molecule FRET studies of polyproline.
Proc. Natl. Acad. Sci. USA 104, 18964–18969. [PDF]
2006
Camenisch, U., Dip, R., Briand Schumacher, S., Schuler, B. & Naegeli, H. (2006)
Recognition of helical kinks by xeroderma pigmentosum group A protein triggers DNA excision repair.
Nature Struct. Mol. Biol. 13, 278–284. [PDF]
2005
Schuler, B., Lipman, E. A., Steinbach, P. J., Kumke, M. & Eaton, W. A. (2005)
Polyproline and the “spectroscopic ruler” revisited with single molecule fluorescence.
Proc. Natl. Acad. Sci. USA 102, 2754-2759. [PDF]
See Highlight in Science 307, 1016, “Editors’ choice”.
Schuler, B. (2005)
Single molecule spectroscopy of protein folding.
ChemPhysChem 6, 1206-1220. [PDF]
2004
Rhoades, E., Cohen, M., Schuler, B. & Haran, G. (2004)
Two-state folding observed in individual protein molecules.
J. Am. Chem. Soc. 126, 14686-14687. [PDF]
2003 and earlier
Lipman, E. A.*, Schuler, B.*, Bakajin, O. & Eaton, W. A. (2003)
Single molecule measurement of protein folding kinetics.
Science 301, 1233-1235. [PDF] [supplement]
Buscaglia, M., Schuler, B., Lapidus, L. J., Eaton, W. A. & Hofrichter, J. (2003)
Kinetics of Intramolecular Contact Formation in a Denatured Protein.
J. Mol. Biol. 332, 9-12. [PDF]
Schuler, B.*, Lipman, E. A.* & Eaton, W. A. (2002)
Probing the free energy surface for protein folding with single molecule fluorescence spectroscopy.
Nature 419, 743-747. [PDF] [supplement]
Schuler, B., Kremer, W., Kalbitzer, H. R. & Jaenicke, R. (2002)
Role of entropy in protein thermostability: Folding kinetics of a hyperthermophilic cold shock protein at high temperature using 19F-NMR.
Biochemistry 41, 11670-11680. [PDF]
Schuler, B. & Pannell, L. K. (2002)
Specific labeling of polypeptides at amino-terminal cysteine residues using Cy5-benzyl thioester.
Bioconjugate Chem. 13, 1039-1043. [PDF]
Kremer, W., Schuler, B., Harrieder, S., Geyer, M., Gronwald, W., Welker, C., Jaenicke, R. & Kalbitzer, H. R. (2001)
Solution NMR structure of the cold-shock protein from the hyperthermophilic bacterium Thermotoga maritima.
Eur. J. Biochem. 268, 2527-2539. [PDF]
Schuler, B., Fürst, F., Osterroth, F., Steinbacher, S., Huber, R. & Seckler, R. (2000)
Plasticity and steric strain in a parallel ß-helix: Directed mutagenesis of the P22 tailspike protein.
Proteins 39, 89-101. [PDF]
Schuler, B. & Seckler, R. (1999)
The isolated P22 tailspike ß-helix domain: Formation of fibrous aggregates from a nonnative intermediate.
J. Biol. Chem. 274, 18589-18596. [PDF]
Schuler, B. & Seckler, R. (1998)
P22 tailspike folding mutants revisited: Effects on the thermodynamic stability of the isolated ß-helix domain.
J. Mol. Biol. 281, 227-234. [PDF]
Miller, S.*, Schuler, B.* & Seckler, R. (1998)
A reversibly unfolding fragment of P22 tailspike protein with native structure: The isolated ß-helix domain.
Biochemistry 37, 9160-9168. [PDF]
Miller, S., Schuler, B. & Seckler, R. (1998)
Phage P22 tailspike protein: Removal of head-binding domain unmasks effects of folding mutations on native-state thermal stability.
Protein Sci. 7, 2223-2232. [PDF]