|
Inhaltsübersicht | Nanomaschinen | Moleküle | Programme | Kurse | Fun | Links |
||
| > |
Selenocysteine Synthase
If you have visited your local health food store or looked closely at the ingredients in your daily multivitamin, you may have noticed that the element selenium is often listed as one of the beneficial supplements. Selenium is a double-edged sword, however. In general, selenium compounds are toxic and have an unpleasant garlicy odor, but in trace amounts, selenium is essential for our health. Selenium atoms are similar to sulfur atoms, with similar properties, except that selenium compounds tend to be more reactive. In a few specialized proteins, this extra reactivity is just what is needed. For instance, by using a selenium atom instead of sulfur, thioredoxin reductase improves its rate of catalysis by 100 times, and formate dehydrogenases act 300 times faster.
Selenocysteine and Selenomethionine
Proteins contain two types of amino acids with sulfur atoms, and both may be made in versions with selenium atoms. Selenomethionine is not made specifically by cells, but it is created occasionally by accident by the normal enzymes that make methionine. This is not a problem, however, since it is similar enough to normal methionine. It is, however, very useful to crystallographers to help with the determination of x-ray crystal structures, since it has more electrons than sulfur and is easily located in x-ray experiments. Selenocysteine, on the other hand, is more reactive than normal cysteine, and it is specifically added to special selenoproteins when it is needed.
Synthesis with Selenium
Quite surprisingly, cells modify their genetic code to add selenocysteine into their proteins. The basic genetic code used by all organisms on Earth specifies twenty amino acids, along with a few stop codons. In order to add a 21st amino acid to this code, cells reinterpret UGA stop codons in special cases. But this causes a problem: how does a cell know when to read UGA as "stop" and when to read it as "selenocysteine"? This is done by using a special signal sequence that is located after the UGA codon. In bacteria, this signal is immediately after the UGA codon, in the coding portion of the messenger RNA. In our cells, the signal is much further away at the end of the coding sequence. As shown on the next page, this sequence is recognized by a special elongation factor that delivers the selenocysteine tRNA to the ribosome at just the right moment.
Slippery Selenium
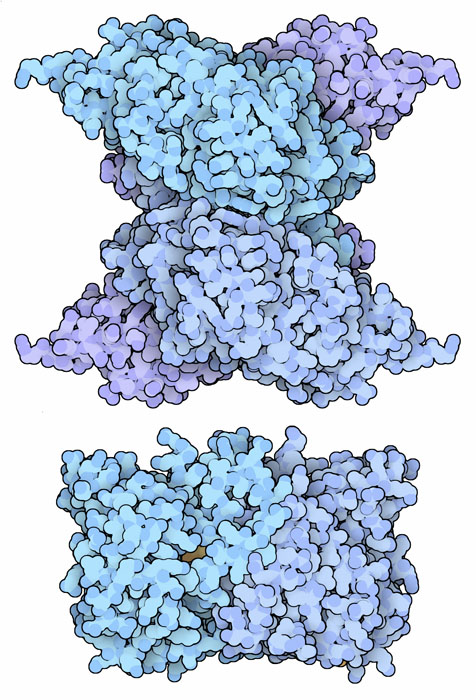
Since selenium and sulfur are so similar, cells must pay careful attention to how selenocysteine is attached to tRNA. They have to make sure that selenocysteine is not added to normal cysteine tRNA, otherwise selenocysteine might be added mistakenly in the wrong place. To solve this problem, selenocysteine tRNA is made in several steps. First, a serine is attached to the selenocysteine tRNA. Then, a second enzyme phosphorylates the serine, making it ready. Finally, the oxygen in the serine is swapped for selenium by the enzyme selenocysteine synthase, shown here from PDB entry 3bc8. The small enzyme selenophosphate synthase, shown at the bottom from PDB entry 2yye, creates an activated form of selenium that is needed in this reaction. By using this stepwise process, none of the tRNA-charging enzymes are ever faced with the delicate task of differentiating cysteine from selenocysteine.
Next: Selecting Selenium
Last changed by: A.Honegger,